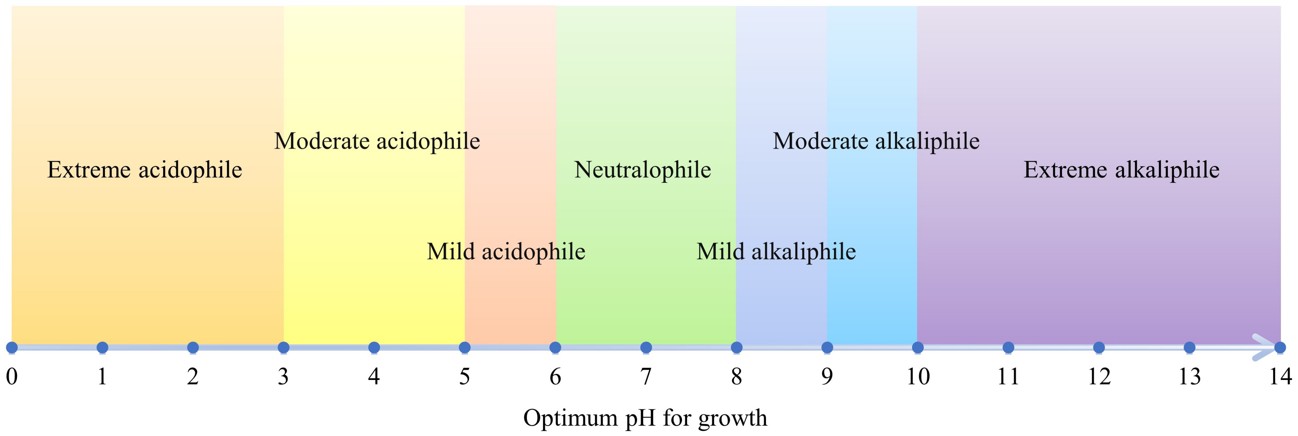
Microorganisms are classified based on their optimal growth pH ranges
The final one-hot encoding matrix for the presence or absence of whole genes
| Optimum pH |
Bacteria |
Gene a |
Gene b |
... |
Gene 20794 |
| 7.25 |
Genome 1 |
0 |
1 |
. |
1 |
| 8.90 |
Genome 2 |
1 |
0 |
. |
0 |
| 3.40 |
Genome 3 |
1 |
0 |
. |
1 |
| ... |
... |
. |
. |
. |
. |
| 5.75 |
Genome 3476 |
0 |
1 |
. |
1 |
Performance of the XGBoost model on the test set
| Model evaluation |
Value |
| MAE test above avg |
0.477490 |
| MSE test above avg |
0.443375 |
| RMSE test above avg |
0.665864 |
| R² test above avg |
0.350277 |
Performance of the XGBoost model on the validation set
| Model evaluation |
Value |
| MAE validation above avg |
0.492234 |
| MSE validation above avg |
0.481866 |
| RMSE validation above avg |
0.694166 |
| R² validation above avg |
0.416336 |
Calculate the results at different precisions using accuracy as the indicator.
| Allowable error |
Accuracy on the test set |
Accuracy on the validation set |
| 0.5 pH units |
0.654952 |
0.632184 |
| 0.6 pH units |
0.728435 |
0.724138 |
| 0.7 pH units |
0.779553 |
0.781609 |
| 0.8 pH units |
0.837061 |
0.830460 |
| 0.9 pH units |
0.869010 |
0.867816 |
| 1.0 pH units |
0.888179 |
0.893678 |
| 2.0 pH units |
0.984026 |
0.979885 |
SHAP analysis of the top 20 genes with the greatest impact
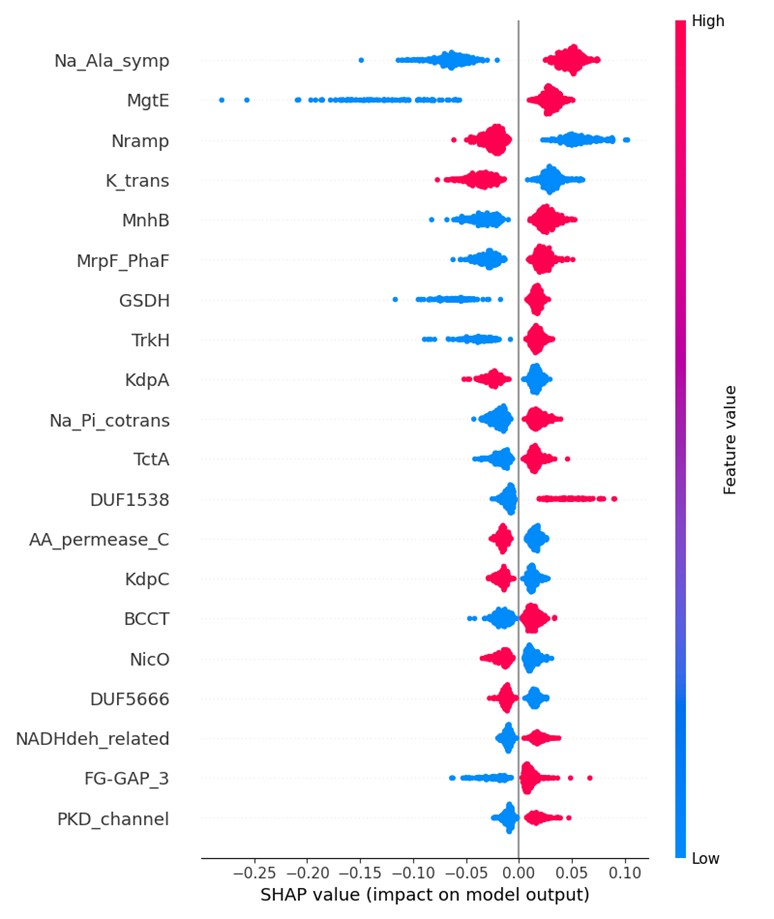
SHAP analysis of the top 20 genes among the 5485 feature genes showed that most of the genes with significant impacts are key genes reported in literature to be involved in the physiological mechanisms of microbial responses to changes in the external pH environment.
For instance, the Na_Ala_symp gene belongs to the sodium/alanine symporter family, and the protein encoded by this gene transports alanine through binding to sodium ions. The MgtE gene is classified as a transmembrane Mg²⁺ transporter, which can transport Mg²⁺ or other divalent cations into cells.
Modules used for project completion
| Name |
Version |
Build |
Channel |
| _libgcc_mutex |
0.1 |
conda_forge |
conda-forge |
| _openmp_mutex |
4.5 |
2_gnu |
conda-forge |
| _py-xgboost-mutex |
2.0 |
cpu_0 |
conda-forge |
| bzip2 |
1.0.8 |
hd590300_5 |
conda-forge |
| ca-certificates |
2024.2.2 |
hbcca054_0 |
conda-forge |
| joblib |
1.3.2 |
pyhd8ed1ab_0 |
conda-forge |
| ld_impl_linux-64 |
2.40 |
h41732ed_0 |
conda-forge |
| libblas |
3.9.0 |
21_linux64_openblas |
conda-forge |
| libcblas |
3.9.0 |
21_linux64_openblas |
conda-forge |
| libffi |
3.4.2 |
h7f98852_5 |
conda-forge |
| libgcc-ng |
13.2.0 |
h807b86a_5 |
conda-forge |
| libgfortran-ng |
13.2.0 |
h69a702a_5 |
conda-forge |
| libgfortran5 |
13.2.0 |
ha4646dd_5 |
conda-forge |
| libgomp |
13.2.0 |
h807b86a_5 |
conda-forge |
| liblapack |
3.9.0 |
21_linux64_openblas |
conda-forge |
| libnsl |
2.0.1 |
hd590300_0 |
conda-forge |
| libopenblas |
0.3.26 |
pthreads_h413a1c8_0 |
conda-forge |
| libsqlite |
3.45.1 |
h2797004_0 |
conda-forge |
| libstdcxx-ng |
13.2.0 |
h7e041cc_5 |
conda-forge |
| libuuid |
2.38.1 |
h0b41bf4_0 |
conda-forge |
| libxcrypt |
4.4.36 |
hd590300_1 |
conda-forge |
| libxgboost |
2.0.3 |
cpu_h6728c87_1 |
conda-forge |
| libzlib |
1.2.13 |
hd590300_5 |
conda-forge |
| ncurses |
6.4 |
h59595ed_2 |
conda-forge |
| numpy |
1.26.4 |
py310hb13e2d6_0 |
conda-forge |
| openssl |
3.2.1 |
hd590300_0 |
conda-forge |
| pandas |
2.2.1 |
py310hcc13569_0 |
conda-forge |
| pip |
24.0 |
pyhd8ed1ab_0 |
conda-forge |
| py-xgboost |
2.0.3 |
cpu_pyh0a621ce_1 |
conda-forge |
| python |
3.10.13 |
hd12c33a_1_cpython |
conda-forge |
| python-dateutil |
2.8.2 |
pyhd8ed1ab_0 |
conda-forge |
| python-tzdata |
2024.1 |
pyhd8ed1ab_0 |
conda-forge |
| python_abi |
3.10 |
4_cp310 |
conda-forge |
| pytz |
2024.1 |
pyhd8ed1ab_0 |
conda-forge |
| readline |
8.2 |
h8228510_1 |
conda-forge |
| scikit-learn |
1.4.1.post1 |
py310h1fdf081_0 |
conda-forge |
| scipy |
1.12.0 |
py310hb13e2d6_2 |
conda-forge |
| setuptools |
69.1.1 |
pyhd8ed1ab_0 |
conda-forge |
| six |
1.16.0 |
pyh6c4a22f_0 |
conda-forge |
| threadpoolctl |
3.3.0 |
pyhc1e730c_0 |
conda-forge |
| tk |
8.6.13 |
noxft_h4845f30_101 |
conda-forge |
| tzdata |
2024a |
h0c530f3_0 |
conda-forge |
| wheel |
0.42.0 |
pyhd8ed1ab_0 |
conda-forge |
| xgboost |
2.0.3 |
cpu_pyhb06c54e_1 |
conda-forge |
| xz |
5.2.6 |
h166bdaf_0 |
conda-forge |


